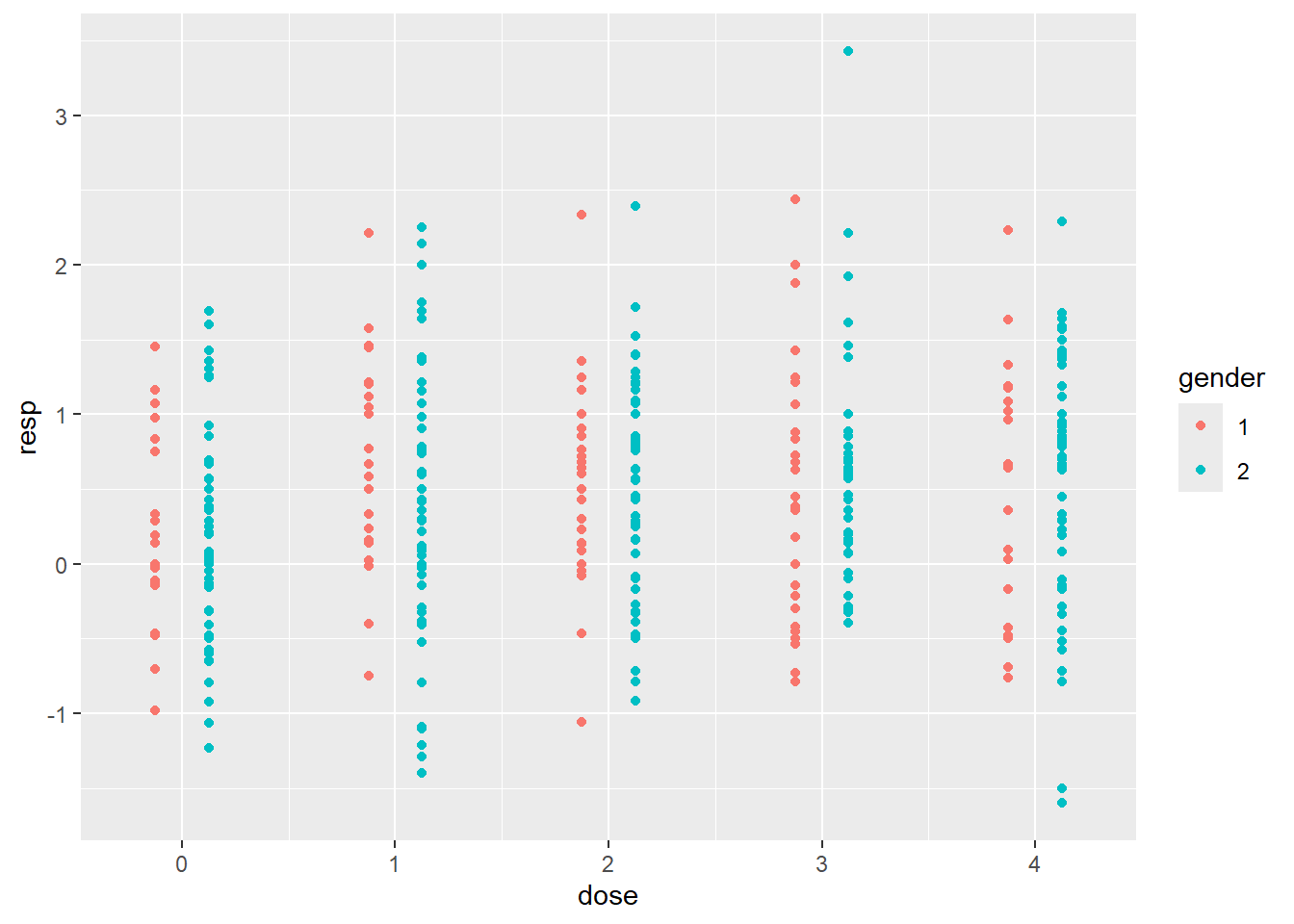
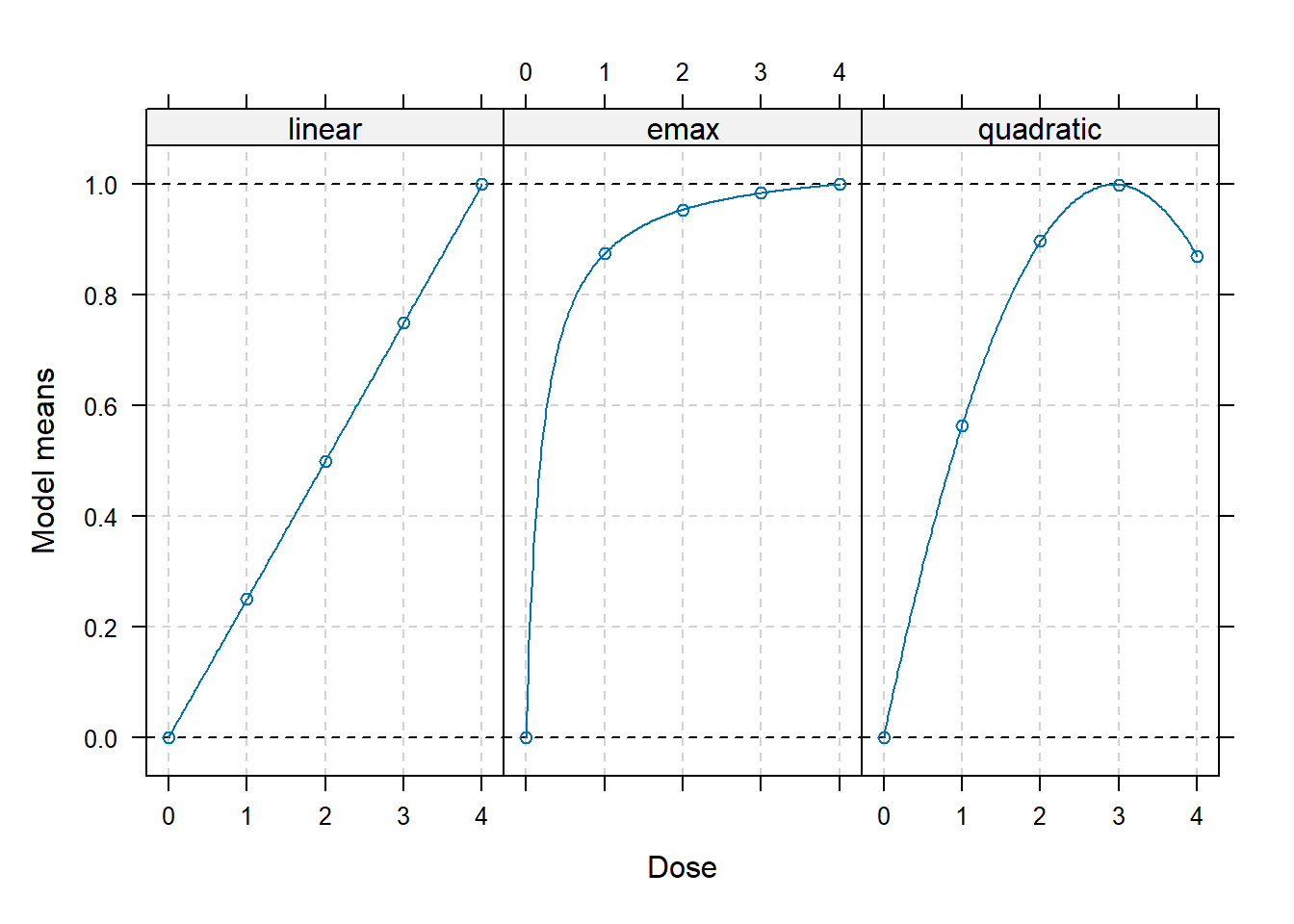
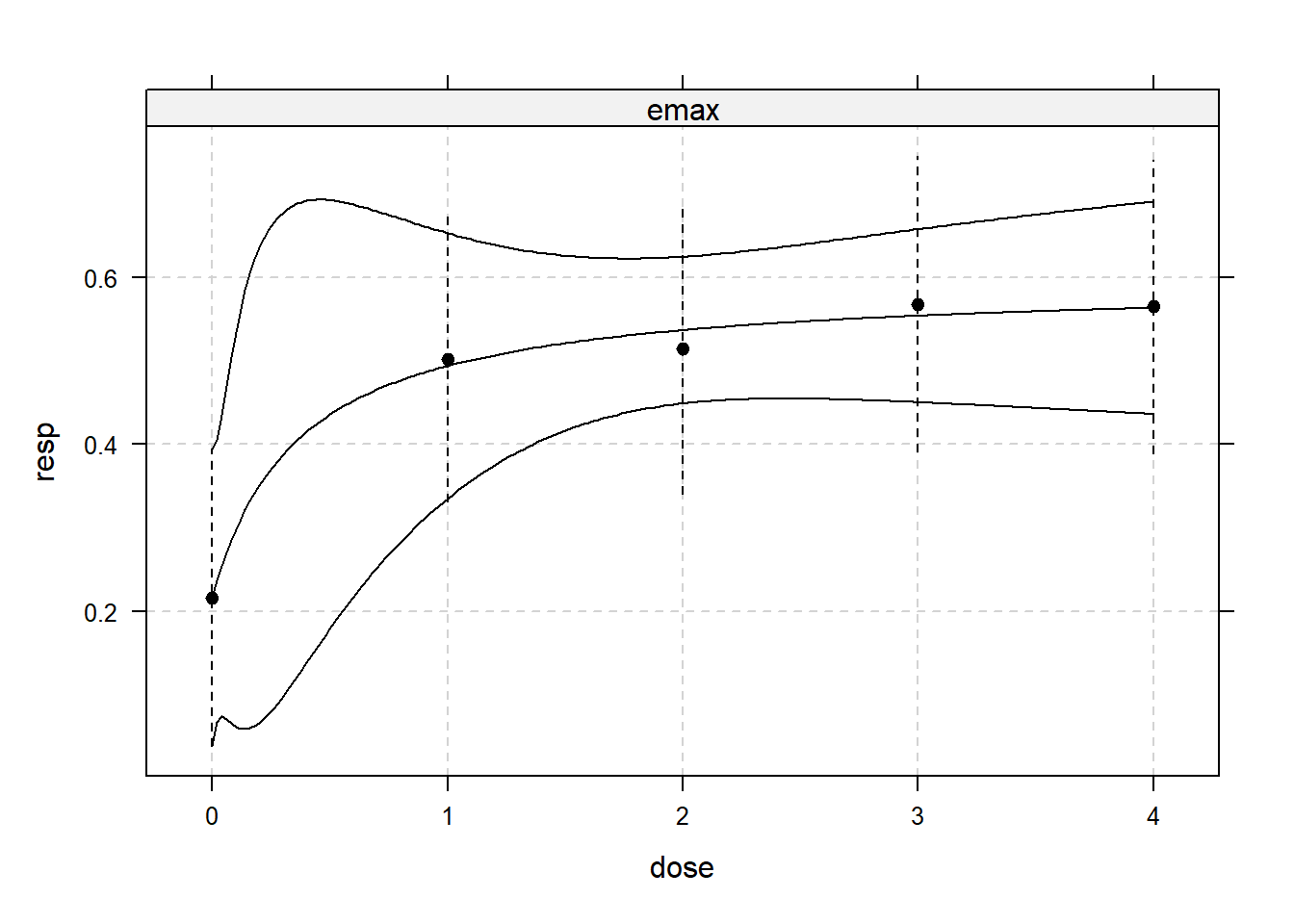
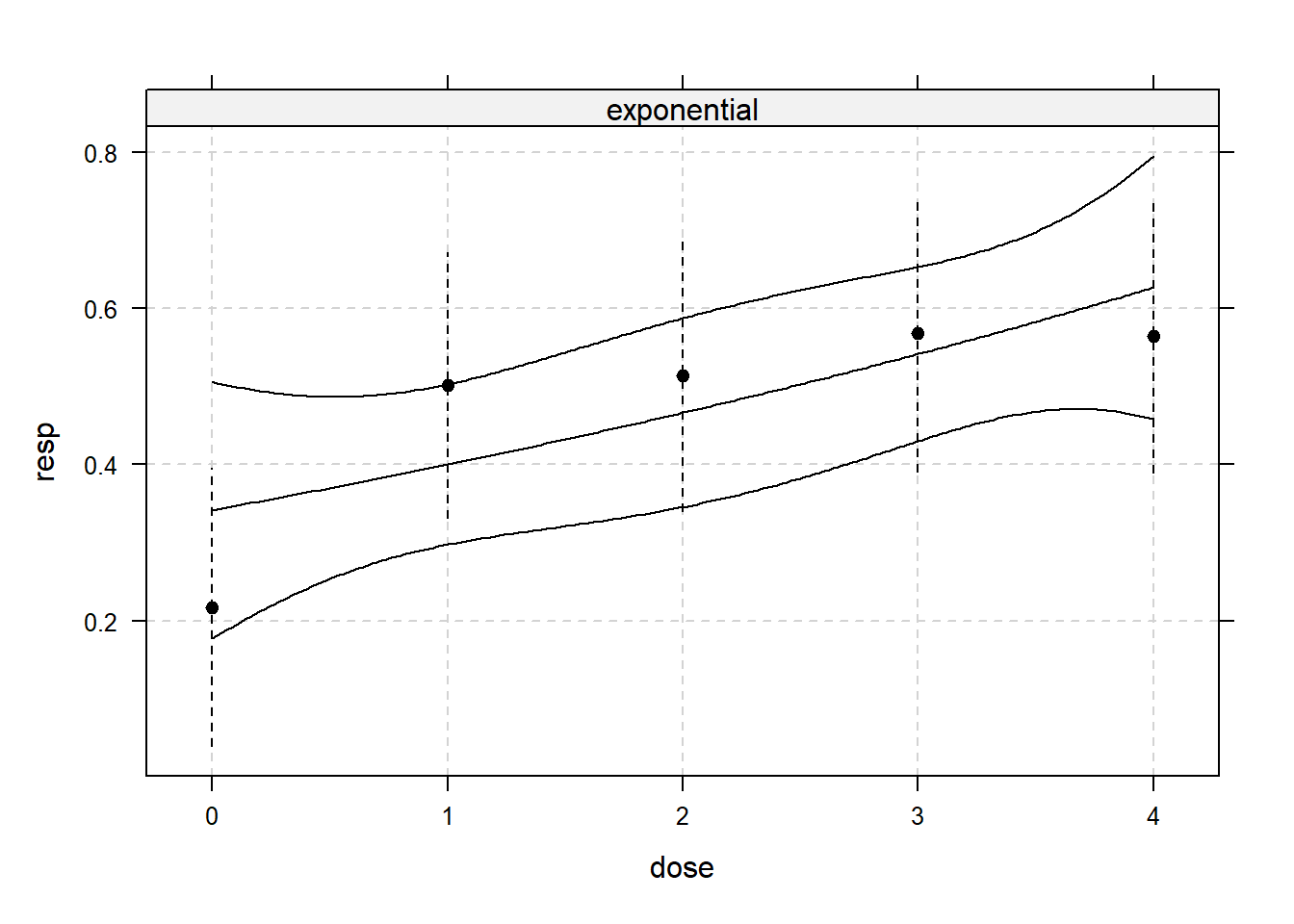
library(DoseFinding)
library(ggplot2)
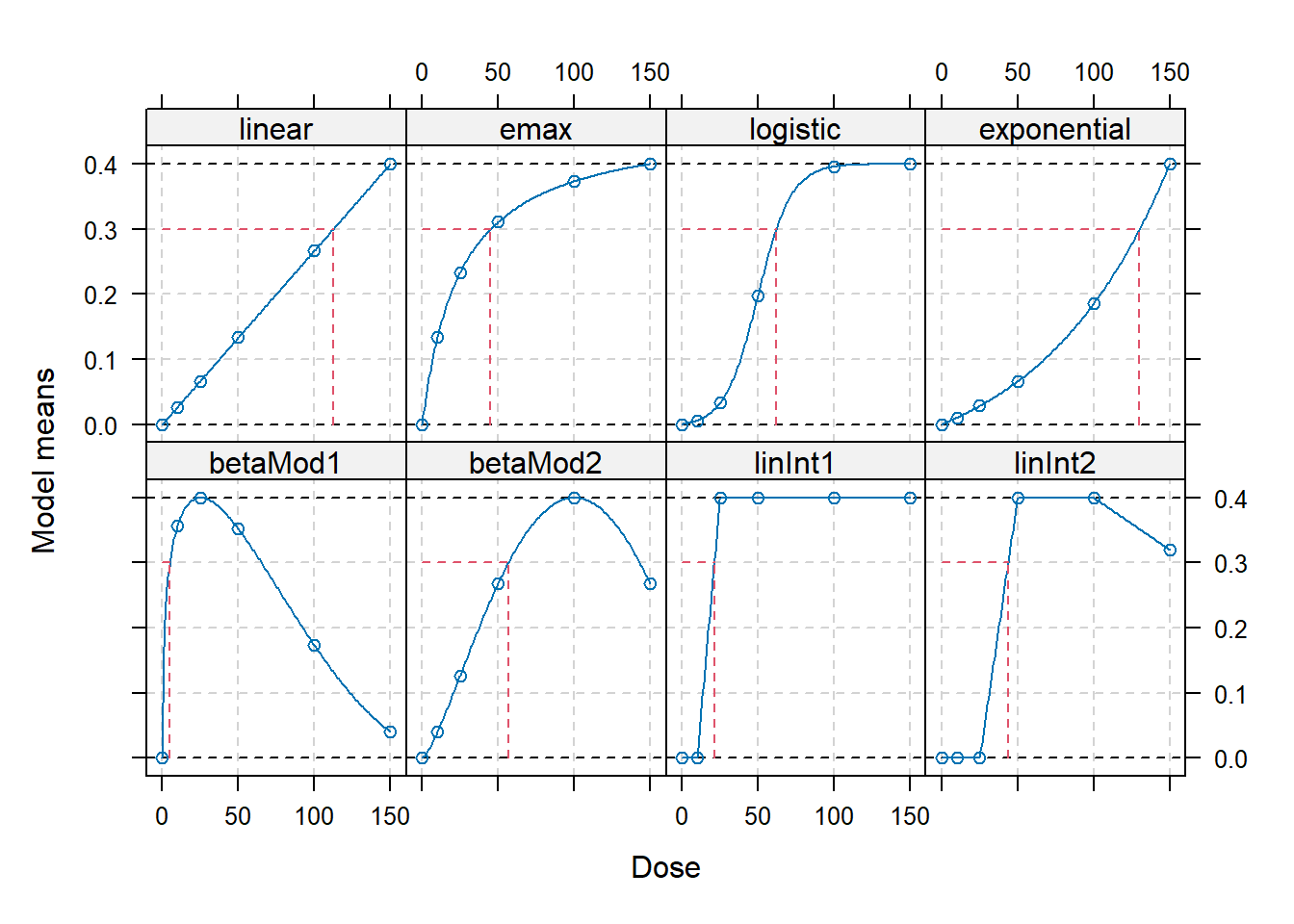
## example for creating a "full-model" candidate set placebo response
## and maxEff already fixed in Mods call
doses <- c(0, 10, 25, 50, 100, 150)
fmodels <- Mods(linear = NULL, emax = 25,
logistic = c(50, 10.88111), exponential = 85,
betaMod = rbind(c(0.33, 2.31), c(1.39, 1.39)),
linInt = rbind(c(0, 1, 1, 1, 1),
c(0, 0, 1, 1, 0.8)),
doses=doses, placEff = 0, maxEff = 0.4,
addArgs=list(scal=200))
## calculate doses giving an improvement of 0.3 over placebo
## Target doses 63
TD(fmodels, Delta=0.3)