Code
library(tidyverse)Zhenglei Gao
January 10, 2025
GLMcat is an R package that encompasses lots of models specified in a similar way: (ratio, cdf, design: parallel or complete).
ratio can be cumulative, sequential, or adjacent for ordinal data, reference for nominal data. Logistic, normal, Cauchy, Student, Gompertz and Gumbel are the commonly used link functions or CDF. Note that Gumbel cdf is the symmetric of the Gompertz cdf.
For parallel design, the effects of the covariates are assumed to be constant across the response categories, whereas for complete design, the covariates effects are different across different response categories. The linear predictios are \(\eta_j=\alpha_j+x'\delta_j\) and \(\eta_j=\alpha_j+x'\delta\) for \(j\) in \((1,...,J-1)\)
There are several type of models for categorical data:
The data was generated by Changjian using SAS Probnorm with Vx=0.25 and Vr=0.15, a=-2.5, b=1.0, ER50=12.18. Model: probit(y) = a + b*logdose + ri;
[1] 0.03539262 0.13270274 0.33703877 0.60741531 0.83291183 0.95143032dattab_new <- read.table(textConnection("Obs rep dose yt y0
1 1 2 0.02021 0
2 2 2 0.02491 0
3 3 2 0.00760 0
4 5 2 0.04466 0
5 6 2 0.00037 0
6 8 2 0.05386 0
7 9 2 0.07205 0
8 10 2 0.01125 0
9 1 4 0.02011 0
10 7 4 0.09058 0
11 8 4 0.06255 0
12 10 8 0.09431 0
13 4 2 0.14060 A
14 7 2 0.22223 A
15 2 4 0.20943 A
16 3 4 0.20959 A
17 4 4 0.17305 A
18 9 4 0.11405 A
19 10 4 0.17668 A
20 1 8 0.12756 A
21 6 8 0.11478 A
22 6 32 0.20602 A
23 5 4 0.26650 B
24 6 4 0.27344 B
25 3 8 0.27021 B
26 5 8 0.30662 B
27 8 8 0.29319 B
28 9 8 0.37300 B
29 6 16 0.36224 B
30 9 16 0.31316 B
31 2 8 0.55845 C
32 4 8 0.44811 C
33 3 16 0.42677 C
34 3 32 0.52315 C
35 7 8 0.67080 D
36 2 16 0.71776 D
37 4 16 0.73038 D
38 5 16 0.64232 D
39 7 16 0.68720 D
40 8 16 0.61088 D
41 5 32 0.72342 D
42 8 32 0.63594 D
43 1 16 0.77171 E
44 10 16 0.74087 E
45 2 32 0.79477 E
46 4 32 0.88546 E
47 7 32 0.78002 E
48 9 32 0.81456 E
49 10 32 0.89465 E
50 1 32 0.96129 F
51 1 64 0.96127 F
52 2 64 0.91687 F
53 3 64 0.97204 F
54 4 64 0.99268 F
55 5 64 0.98935 F
56 6 64 0.96263 F
57 7 64 0.95435 F
58 8 64 0.92081 F
59 9 64 0.91776 F
60 10 64 0.99104 F"),header = TRUE)dattab_new <- dattab_new %>% mutate(yy = as.numeric(plyr::mapvalues(y0,from = c("0","A","B","C","D","E","F"),to = c(0,10,30,50,70,90,100)))/100) %>% mutate(yy2 = as.numeric(plyr::mapvalues(y0,from = c("0","A","B","C","D","E","F"),to = c(0.05,0.18,0.34,0.50,0.66,0.82,0.95))))
ftable(xtabs(~ y0 + dose, data = dattab_new)) ##%>% gt::gt() ## as.data.frame(.) %>% knitr::kable(.,digits = 3) dose 2 4 8 16 32 64
y0
0 8 3 1 0 0 0
A 2 5 2 0 1 0
B 0 2 4 2 0 0
C 0 0 2 1 1 0
D 0 0 1 5 2 0
E 0 0 0 2 5 0
F 0 0 0 0 1 10| y0 | n | meany | meanyy2 |
|---|---|---|---|
| 0 | 12 | 0.042 | 0.05 |
| A | 10 | 0.169 | 0.18 |
| B | 8 | 0.307 | 0.34 |
| C | 4 | 0.489 | 0.50 |
| D | 8 | 0.677 | 0.66 |
| E | 7 | 0.812 | 0.82 |
| F | 11 | 0.958 | 0.95 |
| dose | n | meany | meanyy |
|---|---|---|---|
| 2 | 10 | 0.060 | 0.02 |
| 4 | 10 | 0.160 | 0.11 |
| 8 | 10 | 0.326 | 0.31 |
| 16 | 10 | 0.600 | 0.64 |
| 32 | 10 | 0.722 | 0.75 |
| 64 | 10 | 0.958 | 1.00 |
Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
ℹ Please use `linewidth` instead.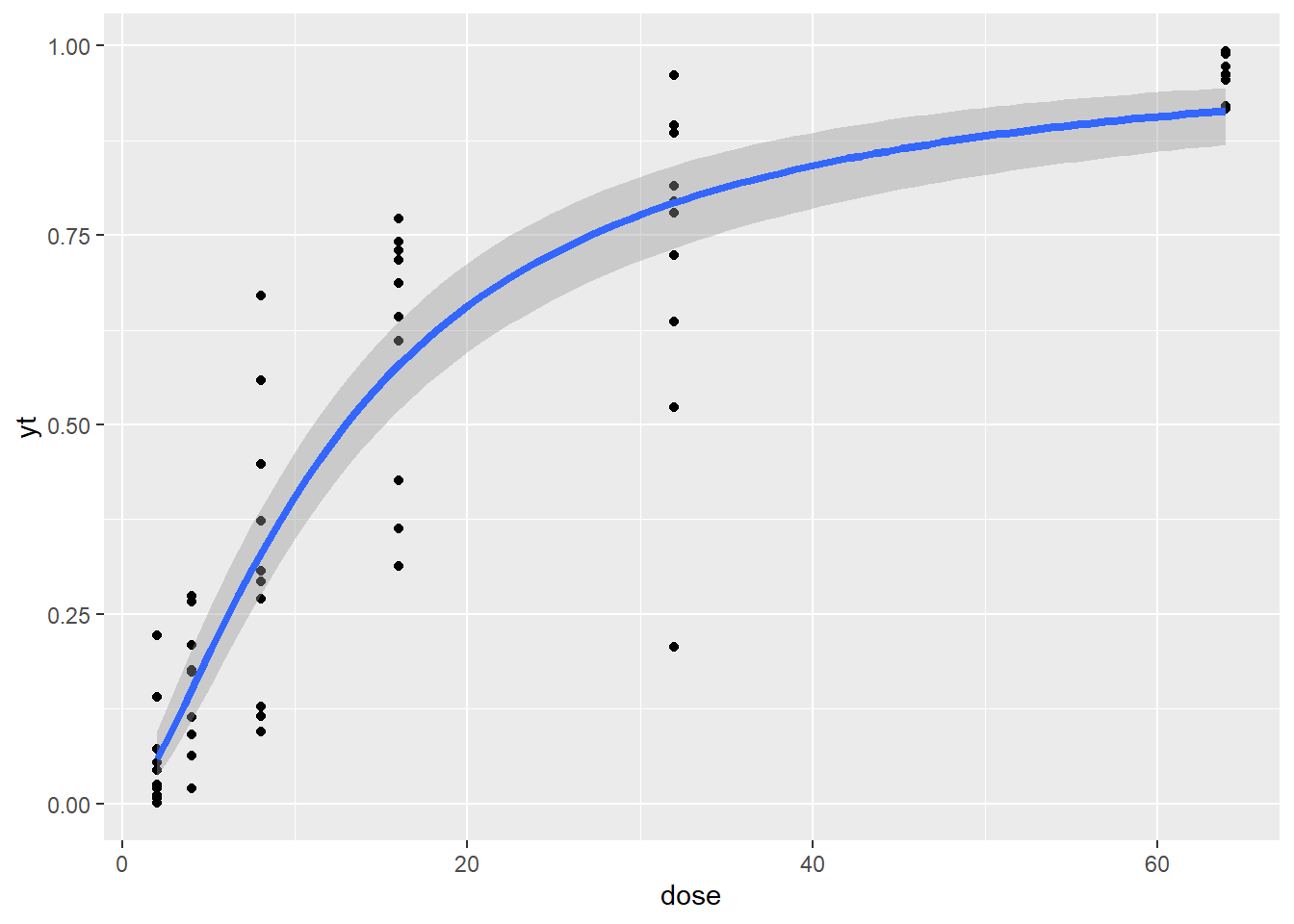
| y0 | dose | 2 | 4 | 8 | 16 | 32 | 64 |
|---|---|---|---|---|---|---|---|
| 0 | 8 | 3 | 1 | 0 | 0 | 0 | |
| A | 2 | 5 | 2 | 0 | 1 | 0 | |
| B | 0 | 2 | 4 | 2 | 0 | 0 | |
| C | 0 | 0 | 2 | 1 | 1 | 0 | |
| D | 0 | 0 | 1 | 5 | 2 | 0 | |
| E | 0 | 0 | 0 | 2 | 5 | 0 | |
| F | 0 | 0 | 0 | 0 | 1 | 10 |
Call:
MASS::polr(formula = y0 ~ log(dose), data = dattab_new, Hess = TRUE)
Coefficients:
Value Std. Error t value
log(dose) 3.465 0.4876 7.106
Intercepts:
Value Std. Error t value
0|A 4.0061 0.8091 4.9513
A|B 6.3415 1.0283 6.1669
B|C 8.1921 1.2395 6.6092
C|D 9.1287 1.3489 6.7677
D|E 10.9709 1.5691 6.9917
E|F 12.9482 1.8234 7.1011
Residual Deviance: 128.7906
AIC: 142.7906 log(dose) 0|A A|B B|C C|D D|E E|F
0.9908438 0.9975973 0.9998653 0.9999875 0.9999963 0.9999997 1.0000000 Value Std. Error t value p value
log(dose) 3.465155 0.4876298 7.106117 1.193530e-12
0|A 4.006148 0.8091031 4.951344 7.370254e-07
A|B 6.341529 1.0283150 6.166913 6.963590e-10
B|C 8.192141 1.2395138 6.609156 3.865169e-11
C|D 9.128696 1.3488680 6.767672 1.308710e-11
D|E 10.970866 1.5691374 6.991654 2.716642e-12
E|F 12.948174 1.8234117 7.101070 1.237950e-12Waiting for profiling to be done... 2.5 % 97.5 %
2.582836 4.504378 2.5 % 97.5 %
log(dose) 31.98141 13.23462 90.41212 OR ci
2.5 % 31.98141 13.23462
97.5 % 31.98141 90.41212 0 A B C D E
1 8.326166e-01 0.1483000019 0.016035611 0.001850927 0.00100702 0.0001635762
2 3.105443e-01 0.5126011687 0.144195119 0.019598385 0.01096824 0.0018025754
3 3.918687e-02 0.2573055374 0.431914336 0.144078094 0.10487831 0.0194406374
4 3.342916e-04 0.0031093320 0.018073161 0.031603441 0.20832988 0.4574198047
5 3.679470e-03 0.0330795765 0.158639125 0.187148646 0.41376767 0.1694853156
6 3.027903e-05 0.0002825179 0.001674417 0.003066947 0.02600507 0.1569497884
F
1 2.628958e-05
2 2.902582e-04
3 3.196213e-03
4 2.811301e-01
5 3.420020e-02
6 8.119910e-01| 0 | A | B | C | D | E | F |
|---|---|---|---|---|---|---|
| 0.833 | 0.148 | 0.016 | 0.002 | 0.001 | 0.000 | 0.000 |
| 0.311 | 0.513 | 0.144 | 0.020 | 0.011 | 0.002 | 0.000 |
| 0.039 | 0.257 | 0.432 | 0.144 | 0.105 | 0.019 | 0.003 |
| 0.000 | 0.003 | 0.018 | 0.032 | 0.208 | 0.457 | 0.281 |
| 0.004 | 0.033 | 0.159 | 0.187 | 0.414 | 0.169 | 0.034 |
| 0.000 | 0.000 | 0.002 | 0.003 | 0.026 | 0.157 | 0.812 |
Warning: package 'GLMcat' was built under R version 4.4.2Warning in glmcat(formula = y0 ~ logdose, ratio = "reference", cdf =
"logistic", : The response variable is defined as an ordered variable. Recall
that the the reference ratio is appropiate for nominal responses, while for
ordinal responses the ratios to use are cumulative, sequential or adjacent.y0 ~ logdose
ratio cdf nobs niter logLik
Model info: reference logistic 60 15 -61.33542
Estimate Std. Error z value Pr(>|z|)
(Intercept) A -2.907 1.309 -2.221 0.026323 *
(Intercept) B -5.398 1.835 -2.942 0.003260 **
(Intercept) C -9.347 3.019 -3.096 0.001958 **
(Intercept) D -10.923 3.065 -3.564 0.000366 ***
(Intercept) E -17.421 4.838 -3.600 0.000318 ***
(Intercept) F -118.572 109742.484 -0.001 0.999138
logdose A 2.179 0.993 2.194 0.028225 *
logdose B 3.415 1.177 2.901 0.003725 **
logdose C 4.803 1.504 3.194 0.001404 **
logdose D 5.633 1.499 3.758 0.000172 ***
logdose E 7.692 1.895 4.058 4.94e-05 ***
logdose F 36.404 31664.987 0.001 0.999083
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 A B C D E
[1,] 1.904456e-01 3.716346e-02 1.873707e-03 6.891895e-04 4.324730e-06
[2,] 4.075170e-01 1.873256e-01 2.472185e-02 1.616443e-02 4.225492e-04
[3,] 3.188563e-01 3.452649e-01 1.192710e-01 1.386298e-01 1.509628e-02
[4,] 5.226961e-03 3.140645e-02 7.433584e-02 2.730262e-01 5.159543e-01
[5,] 7.795418e-02 1.988396e-01 1.797976e-01 3.714905e-01 1.685225e-01
[6,] 2.602306e-12 3.683274e-11 2.281981e-10 1.489911e-09 1.172908e-08
F 0
[1,] 2.238505e-41 7.698238e-01
[2,] 9.621081e-31 3.638486e-01
[3,] 1.512043e-20 6.288173e-02
[4,] 1.000000e-01 5.029071e-05
[5,] 7.425049e-11 3.395654e-03
[6,] 1.000000e+00 5.551115e-15| A | B | C | D | E | F | 0 |
|---|---|---|---|---|---|---|
| 0.190 | 0.037 | 0.002 | 0.001 | 0.000 | 0.0 | 0.770 |
| 0.408 | 0.187 | 0.025 | 0.016 | 0.000 | 0.0 | 0.364 |
| 0.319 | 0.345 | 0.119 | 0.139 | 0.015 | 0.0 | 0.063 |
| 0.005 | 0.031 | 0.074 | 0.273 | 0.516 | 0.1 | 0.000 |
| 0.078 | 0.199 | 0.180 | 0.371 | 0.169 | 0.0 | 0.003 |
| 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 1.0 | 0.000 |
y0 ~ logdose
ratio cdf nobs niter logLik
Model info: cumulative logistic 60 9 -64.3953
Estimate Std. Error z value Pr(>|z|)
(Intercept) 0 4.0061 0.8142 4.920 8.64e-07 ***
(Intercept) A 6.3415 1.0419 6.086 1.16e-09 ***
(Intercept) B 8.1920 1.2583 6.510 7.50e-11 ***
(Intercept) C 9.1286 1.3693 6.666 2.62e-11 ***
(Intercept) D 10.9707 1.6005 6.854 7.16e-12 ***
(Intercept) E 12.9479 1.8194 7.117 1.11e-12 ***
logdose -3.4651 0.4912 -7.054 1.74e-12 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 0 A B C D E
[1,] 8.326186e-01 0.1482975531 0.016035767 0.001851046 0.001007102 0.000163596
[2,] 3.105557e-01 0.5125923926 0.144191357 0.019598856 0.010968693 0.001802725
[3,] 3.919032e-02 0.2573138937 0.431903119 0.144076171 0.104878455 0.019441472
[4,] 3.343480e-04 0.0031097342 0.018074387 0.031605029 0.208331116 0.457408504
[5,] 3.679947e-03 0.0330824671 0.158642283 0.187148395 0.413758936 0.169485389
[6,] 3.028531e-05 0.0002825654 0.001674598 0.003067232 0.026006412 0.156948672
F
[1,] 2.629451e-05
[2,] 2.903015e-04
[3,] 3.196565e-03
[4,] 2.811369e-01
[5,] 3.420258e-02
[6,] 8.119902e-01| 0 | A | B | C | D | E | F |
|---|---|---|---|---|---|---|
| 0.833 | 0.148 | 0.016 | 0.002 | 0.001 | 0.000 | 0.000 |
| 0.311 | 0.513 | 0.144 | 0.020 | 0.011 | 0.002 | 0.000 |
| 0.039 | 0.257 | 0.432 | 0.144 | 0.105 | 0.019 | 0.003 |
| 0.000 | 0.003 | 0.018 | 0.032 | 0.208 | 0.457 | 0.281 |
| 0.004 | 0.033 | 0.159 | 0.187 | 0.414 | 0.169 | 0.034 |
| 0.000 | 0.000 | 0.002 | 0.003 | 0.026 | 0.157 | 0.812 |
Call:
MASS::polr(formula = y0 ~ 1, data = dattab_new, Hess = T)
No coefficients
Intercepts:
0|A A|B B|C C|D D|E
-1.386294e+00 -5.465462e-01 -1.793225e-06 2.682586e-01 8.472940e-01
E|F
1.493920e+00
Residual Deviance: 228.003
AIC: 240.003 0|A A|B B|C C|D D|E E|F
0.2000001 0.3666661 0.4999996 0.5666653 0.6999992 0.8166659 Length Class Mode
3 formula call
Call: glm(formula = I(as.numeric(y0) >= 2) ~ log(dose), family = "binomial",
data = dattab_new)
Coefficients:
(Intercept) log(dose)
-3.305 2.874
Degrees of Freedom: 59 Total (i.e. Null); 58 Residual
Null Deviance: 60.05
Residual Deviance: 29.19 AIC: 33.19
Call: glm(formula = I(as.numeric(y0) >= 3) ~ log(dose), family = "binomial",
data = dattab_new)
Coefficients:
(Intercept) log(dose)
-5.089 2.726
Degrees of Freedom: 59 Total (i.e. Null); 58 Residual
Null Deviance: 78.86
Residual Deviance: 33.83 AIC: 37.83
Call: glm(formula = I(as.numeric(y0) >= 4) ~ log(dose), family = "binomial",
data = dattab_new)
Coefficients:
(Intercept) log(dose)
-7.440 3.067
Degrees of Freedom: 59 Total (i.e. Null); 58 Residual
Null Deviance: 83.18
Residual Deviance: 30.69 AIC: 34.69getEC50 <- function(mod){
if("glm" %in% class(mod)){
ec50 <- MASS::dose.p(mod,p=0.5)
se <- attr(ec50,"SE")
xmin <- as.numeric(ec50 - 1.96*se)
xmax <- as.numeric(ec50 + 1.96*se)
res <- data.frame(EC50=exp(as.numeric(ec50)),
lower=exp(xmin),upper=exp(xmax),se=se)
}else{
if("glmmPQL" %in% class(mod)){
res1 <- dose.p.glmmPQL(mod,cf=1:2,p=0.5)
se <- attr(res1,"SE")
res <- data.frame(EC50=exp(res1[1]),lower=exp(res1[1]-1.96*se),upper=exp(res1[1]+1.96*se),se=se)
}else{
res1 <- ED(mod,50,interval="delta")
res <- data.frame(EC50=res1[1,"Estimate"],lower=res1[1,"Lower"],upper=res1[1,"Upper"],se=res1[1,"Std. Error"])
}
}
return(res)
}
dose.p.glmmPQL <- function(obj,cf=1:2,p=0.5){
eta <- obj$family$linkfun(p)
b <- obj$coefficients$fixed[cf]
x.p <- (eta - b[1L])/b[2L]
names(x.p) <- paste("p = ", format(p), ":", sep = "")
pd <- -cbind(1, x.p)/b[2L]
SE <- sqrt(((pd %*% vcov(obj)[cf, cf]) * pd) %*% c(1, 1))
res <- structure(x.p, SE = SE, p = p)
class(res) <- "glm.dose"
res
}
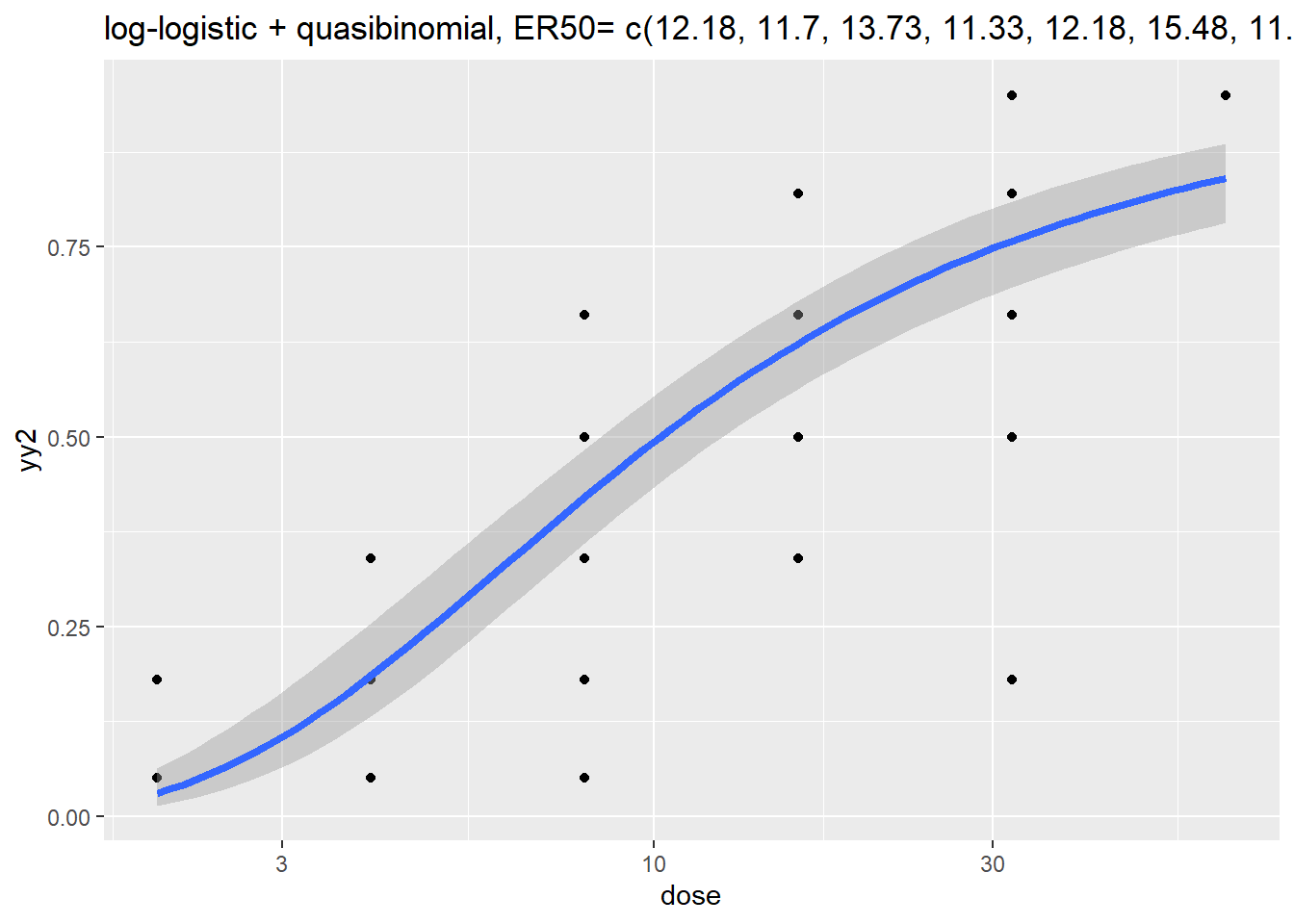
Call:
glm(formula = yy2 ~ log(dose), family = quasibinomial, data = dattab_new)
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -3.5064 0.3132 -11.19 4.02e-16 ***
log(dose) 1.3832 0.1171 11.81 < 2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for quasibinomial family taken to be 0.1163524)
Null deviance: 31.8277 on 59 degrees of freedom
Residual deviance: 6.3595 on 58 degrees of freedom
AIC: NA
Number of Fisher Scoring iterations: 5(Intercept)
12.617 EC50 lower upper se
p = 0.5: 12.617 10.77038 14.78023 0.08073738| y0 | n | meany | meanyy2 | meanEst |
|---|---|---|---|---|
| 0 | 12 | 0.042 | 0.05 | 0.120 |
| A | 10 | 0.169 | 0.18 | 0.247 |
| B | 8 | 0.307 | 0.34 | 0.361 |
| C | 4 | 0.489 | 0.50 | 0.515 |
| D | 8 | 0.677 | 0.66 | 0.603 |
| E | 7 | 0.812 | 0.82 | 0.726 |
| F | 11 | 0.958 | 0.95 | 0.893 |
(Intercept)
12.61695 Linear mixed-effects model fit by maximum likelihood
Data: dattab_new
AIC BIC logLik
NA NA NA
Random effects:
Formula: ~1 | rep
(Intercept) Residual
StdDev: 0.2256918 0.3155037
Variance function:
Structure: fixed weights
Formula: ~invwt
Fixed effects: yy2 ~ log(dose)
Value Std.Error DF t-value p-value
(Intercept) -3.517527 0.3039553 49 -11.57251 0
log(dose) 1.387562 0.1103306 49 12.57640 0
Correlation:
(Intr)
log(dose) -0.907
Standardized Within-Group Residuals:
Min Q1 Med Q3 Max
-3.9592347 -0.3047297 0.1382668 0.4944066 1.9353183
Number of Observations: 60
Number of Groups: 10 EC50 lower upper se
p = 0.5: 12.61695 10.56476 15.06777 0.09057011| y0 | n | meany | meanyy2 | meanEst | meanEstR |
|---|---|---|---|---|---|
| 0 | 12 | 0.042 | 0.05 | 0.120 | 0.120 |
| A | 10 | 0.169 | 0.18 | 0.247 | 0.240 |
| B | 8 | 0.307 | 0.34 | 0.361 | 0.342 |
| C | 4 | 0.489 | 0.50 | 0.515 | 0.517 |
| D | 8 | 0.677 | 0.66 | 0.603 | 0.620 |
| E | 7 | 0.812 | 0.82 | 0.726 | 0.736 |
| F | 11 | 0.958 | 0.95 | 0.893 | 0.894 |
Note that glmer and glmmPQL (based on lme from the nlme pacakge) differs in terms of parameter estimation algorithm and nlme is not optimized for dealing with crossed random effects, which are associated with a sparse design matrix. See more in the book from Pinheiro & Bates.[^1]
Model fitted: Log-logistic (ED50 as parameter) with lower limit at 0 and upper limit at 1 (2 parms)
Parameter estimates:
Estimate Std. Error t-value p-value
b:(Intercept) -1.33865 0.13595 -9.8465 5.487e-14 ***
e:(Intercept) 12.74391 1.03919 12.2633 < 2.2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error:
0.1446638 (58 degrees of freedom)
Estimated effective doses
Estimate Std. Error Lower Upper
e:1:50 12.7439 1.0392 10.6637 14.8241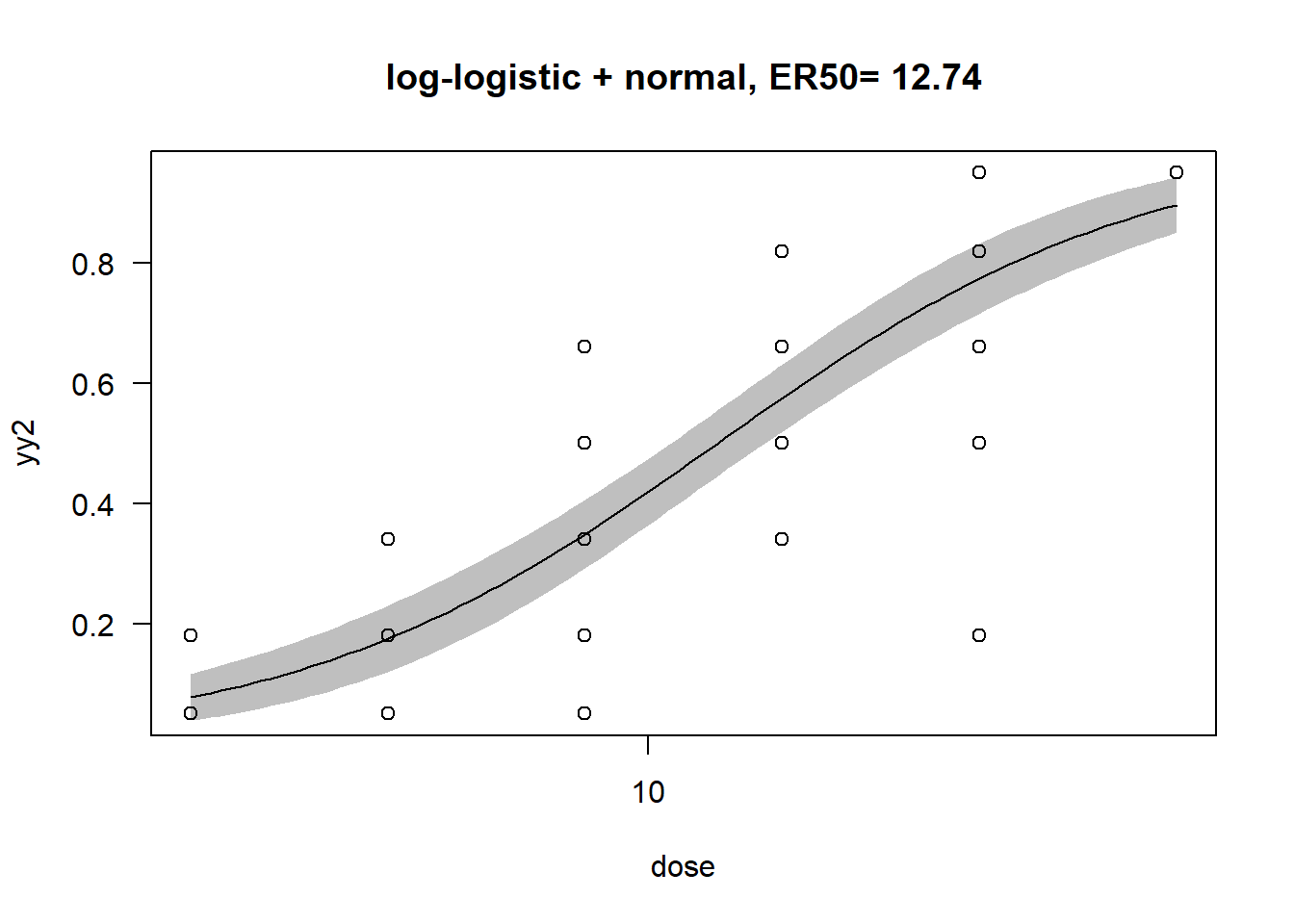
`summarise()` has grouped output by 'y0'. You can override using the `.groups`
argument.dat_ord <- dat_ord %>% mutate(across(c(`0`,`A`,`B`,`C`,`D`,`E`,`F`),~ifelse(is.na(.),0,.))) %>% mutate(total=rowSums(across(c(`0`,`A`,`B`,`C`,`D`,`E`,`F`))))
mod_ord <- drmOrdinal(levels = unique(dattab_new$y0), weights="total",dose = "dose", data = dat_ord, fct = LL.2())
plot(mod_ord) # uses ggplot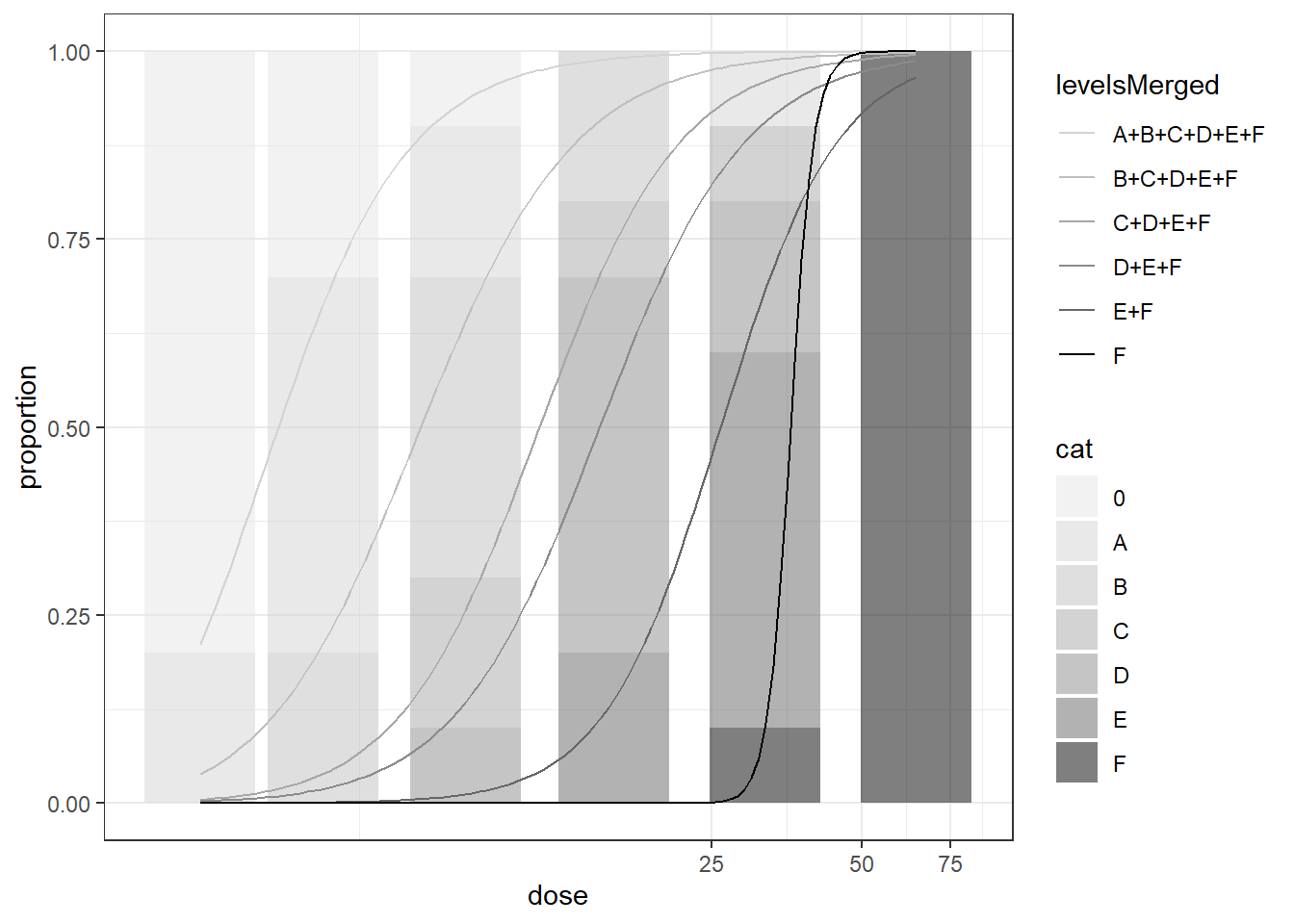
BMD BMDL
A+B+C+D+E+F 3.158208 2.298134
B+C+D+E+F 6.467922 4.757317
C+D+E+F 11.313719 8.507258
D+E+F 14.947346 11.193140
E+F 26.084222 20.202286
F 35.967446 -1.757891 Age Level
Min. : 6.00 Not.severe :100
1st Qu.: 8.50 Severe.1 : 42
Median :10.50 Severe.2 : 41
Mean :10.96 Very.severe: 40
3rd Qu.:12.50
Max. :14.50 Age Level
1 6 Not.severe
2 6 Not.severe
3 6 Not.severe
4 6 Not.severe
5 6 Not.severe
6 6 Not.severeLevel ~ Age
ratio cdf nobs niter logLik
Model info: reference logistic 223 5 -277.1345
Estimate Std. Error z value Pr(>|z|)
(Intercept) Not.severe -2.45444 0.84559 -2.903 0.0037 **
(Intercept) Severe.1 -0.55464 0.89101 -0.622 0.5336
(Intercept) Severe.2 -1.12464 0.91651 -1.227 0.2198
Age Not.severe 0.30999 0.07804 3.972 7.13e-05 ***
Age Severe.1 0.05997 0.08582 0.699 0.4847
Age Severe.2 0.11228 0.08684 1.293 0.1960
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Not.severe Severe.1 Severe.2 Very.severe
[1,] 0.5392049 0.1583321 0.1721826 0.1302804
[2,] 0.1832207 0.2732519 0.2115046 0.3320229
[3,] 0.2996414 0.2391879 0.2110034 0.2501674 Age Level
216 12.5 Very.severe
3 6.0 Not.severe
192 8.5 Very.severe
Attaching package: 'drcData'The following objects are masked from 'package:drc':
auxins, nasturtium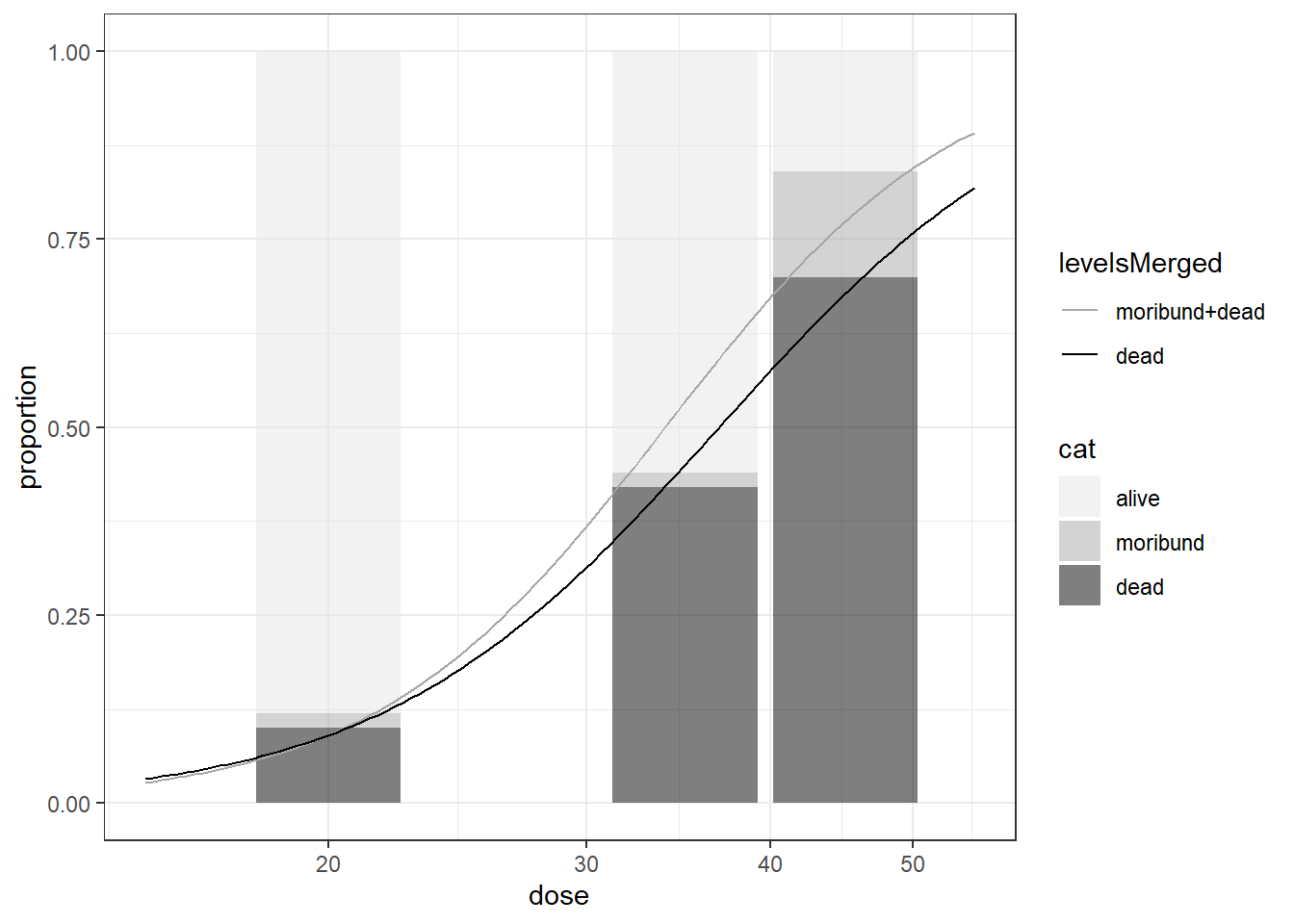
BMD BMDL
moribund+dead 20.50104 17.11484
dead 20.59249 16.69573limited information and pertaintially unequal intervals, which requires a clear understanding of its subtleties and complexities, and careful attentions when implementing regression modelling.
Proportional Odds Model and Continuation Ratio Model, are standard regression models looking at ordered categories and transitions between them
Proportional Odds Model: Coefficient (slopes) remains constant across all categories.
Continuation Ratio Model: Model cumulative odds ratios.
Adjacent Categories Model: useful when neighboring categories influence each other.
[^1] (Pinheiro & Bates, “Mixed-Effects Models in S and S-PLUS”, Springer, 2000, pp. 163. )